Molecular Descriptors Family on Structure Activity Relationships
2. Insecticidal Activity of Neonicotinoid Compounds
Sorana BOLBOACĂ, Lorentz JÄNTSCHI
Iuliu Haţieganu University of Medicine and Pharmacy, http://sorana.academicdirect.ro
Technical University of Cluj-Napoca, Romania, http://lori.academicdirect.org
Abstract
The neonicotinoids are the newest major class of insecticides modeled after the basic nicotine molecule having improved insecticide activity and generally low toxicity. The insecticidal activities of neonicotinoids were previous studied using 3D and standard partial least squares regression models. The paper describes the ability of the MDF SAR methodology in prediction of insecticidal activities of neonicotinoid compounds. The best MDF SAR bi-varied model was validated on training and test sets and its ability on prediction of insecticidal activity was compared with previous reported models. Even if the MDF SAR methodology is complex and time consuming the results worth the effort because they are statistical significant better then previous reported results.
Keywords
Molecular Descriptor Family (MDF), Structure Activity Relationships (SAR), Neonicotinoid Compounds, Insecticidal Activity
Introduction
The neonicotinoids are the newest major class of insecticides modeled after the basic nicotine molecule with improved insecticide activity and generally low toxicity [[1]]. Some benefits of using the neonicotinoid compounds are ([[2]]):
· safer to humans and environmentally friendly vs. older products;
· used as seed coatings at very low amounts per acre;
· are water soluble and highly systemic in seeds and seedlings;
· targeted application (kernel coating) increases efficacy;
· precise application helps minimize environmental effects;
· easy to apply and requires no application equipment;
· reduces storage and transport effort;
· do not have offensive odors;
· are cost competitive.
The product name of the registered neonicotinoid insecticides are [[3]]: Intruder (active ingredient: acetamiprid), Admire (imidacloprid), Provado (imidacloprid), Actara (thiamethoxam), Centric (thiamethoxam), and Platinum (thiamethoxam).
The insecticidal activities of neonicotinoid compounds were previous studied [[4]] using a 3D and standard partial least squares regression (PLS) models. The insecticidal activity of neonicotinoid compounds defined as the logarithm of the reciprocal value of binding constant Ki against the nicotinic acetylcholine (Ach) receptor derived from the head of honeybee. The previous published standard PLS analysis has the following multiple linear regression equation:
n = 8, A = 3, r2 = 0.913, Q2 = 0.800, Log (1/Ki) = 2.045·C1j+1.935·C2j
+1.697·C3j+1.765· C4j+0.695· C5j+0.519· C6j-0.610· C7j+0.254· C8j (1)
where n is the sample size, A is the number of components, r2 is the squared correlation coefficient, C is the Carbo similarity index, and Q2 value is defined as follows:
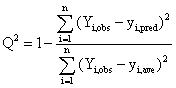 (2)
(2)
where yi,obs and yi,pred are the observed and predicted activity for sample i, respectively, yave is the averaged activity and n is the sample size. The cross-validation results indicate that three is the optimal number of components (see table 1).
Table 1. The 3-way PLS analysis results
|
Component |
1 |
2 |
3 |
4 |
5 |
|
Q2 |
0.597 |
0.791 |
0.855 |
0.803 |
0.829 |
The aim of the research was to estimate the insecticidal activity of neonicotinoid compounds based on an original molecular descriptors family on structure activity relationships method and to estimate the predictive ability by comparing the results with previous reported 3D-QSAR model.
Material and Method
Eight neonicotinoid compounds with insecticidal activity were reported previously as data set for a 3D-QSAR study [4] and were investigate here. The neonicotinoids planar structures and insecticidal activities are in figure 1.
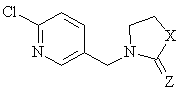
|
Molecule name |
X |
Z |
Insecticidal Activity |
|
na1 |
NH |
CH-NO2 |
7.67 |
|
na2 |
O |
CH-NO2 |
6.51 |
|
na3 |
S |
CH-NO2 |
6.94 |
|
na4 |
C |
CH-NO2 |
6.45 |
|
na5 |
NH |
CH-CN |
4.75 |
|
na6 |
NH |
N-CN |
5.16 |
|
na7 |
N-CH3 |
N-NO2 |
3.79 |
|
na8 |
NH |
N-NO2 |
5.81 |
Figure 1. Eight neonicotinoids acting as insecticides
The methodology of MDF SAR modeling of insecticidal activity of neonicotinoid compounds is [[5]]:
· Sketch of the neonicotinoid compounds;
· Create the neonicotinoid compounds activity file;
· Generate the neonicotinoid compounds MDF members;
· Find the insecticidal activity SAR models;
· Validate and compare with previous reported results the MDF SAR models;
· Analyze the selected MDF SAR model.
Results
The calculated values for the mono and the best bi-varied MDF SAR models and previously reported estimated values (3-way PLS model 3W_PLS, and standard PLS model S_PLS) are in table 2.
Table 2. MDF SAR models and estimated insecticidal activity
|
|
Mono-varied SAR |
Bi-varied SAR |
Tri-varied SAR |
||||
|
19.15-77.49∙iIDrSMg |
43.34-2.21∙ImMdsEg+3.74∙lIMMFQt |
Previously reported [4] |
|||||
|
iIDrSMg |
MDF SAR |
ImMdsEg |
lIMMFQt |
MDF SAR |
3W_PLS |
S_PLS |
|
|
na1 |
0.1574 |
6.961 |
6.135 |
-5.926 |
7.632 |
6.94 |
6.94 |
|
na2 |
0.1531 |
7.293 |
6.951 |
-5.742 |
6.517 |
6.75 |
6.9 |
|
na3 |
0.1566 |
7.023 |
5.665 |
-6.373 |
6.995 |
6.36 |
6.78 |
|
na4 |
0.1662 |
6.275 |
5.667 |
-6.512 |
6.470 |
7.02 |
6.93 |
|
na5 |
0.1852 |
4.804 |
6.855 |
-6.264 |
4.777 |
4.77 |
4.85 |
|
na6 |
0.1843 |
4.877 |
6.940 |
-6.111 |
5.162 |
5.18 |
5.16 |
|
na7 |
0.1938 |
4.140 |
6.174 |
-6.931 |
3.785 |
3.3 |
3.91 |
|
na8 |
0.1736 |
5.706 |
5.985 |
-6.518 |
5.746 |
6.15 |
5.67 |
Mono-varied MDF SAR model has the following statistics:
r = 0.937; r2 = 0.878; r2adj = 0.858 r2cv = 0.756;
F = 43; p = 0.06% (3)
and bi-varied MDF SAR model has the following statistics:
r = 0.999; r2 = 0.999; r2adj = 0.998 r2cv = 0.998;
F = 2864; p = 2·10-6 % (4)
where r is the correlation coefficient, r2 is the r-square, r2adj is the adjusted r2, r2cv is the cross-validation leave one out score, F is the statistical parameter of Fisher test, and p is the probability of wrong associated with F test.
The cross-validation leave one out procedure predicted values for both mono- and bi-varied MDF SAR models are in table 3.
Table 3. Predicted values of insecticidal activity
|
|
na1 |
na2 |
na3 |
na4 |
na5 |
na6 |
na7 |
na8 |
|
Mono-varied model |
6.734 |
7.672 |
7.052 |
6.247 |
4.821 |
4.794 |
4.408 |
5.69 |
|
Bi-varied model |
7.606 |
6.522 |
7.024 |
6.479 |
4.788 |
5.162 |
3.775 |
5.73 |
The plot of mono-varied MDF SAR model values for insecticidal activity of neonicotinoid compounds is in figure 2, and for bi-varied MDF SAR model is in figure 3.

Figure 2. Plot of insecticidal activity vs. mono-varied MDF SAR model

Figure 3. Plot of insecticidal activity vs. bi-varied MDF SAR model
In order to validate the models, the set was split into the training and the test sets and the MLR models were rebuilt. The number of molecules from training set was chouse to be equal with four. The experiment was run for three times just for the bi-varied MDF SAR model. The results are in table 4.
Table 4. Training vs. test sets results
|
|
Molecules in training set |
Model |
r2 |
F, pF(%) |
||
|
Train |
Test |
Train |
Test |
|||
|
1 |
na3,na7,na8,na2 |
42.7-2.17·ImMdsEg+3.69·lIMMFQt |
.999 |
.999 |
490, 3.2 |
382, 3.6 |
|
2 |
na8,na5,na4,na7 |
43.4-2.23·ImMdsEg+3.72·lIMMFQt |
.999 |
.998 |
452, 3.3 |
228, 4.7 |
|
3 |
na4,na7,na6,na8 |
43.3-2.21·ImMdsEg+3.73·lIMMFQt |
.999 |
.999 |
485, 3.2 |
339, 3.8 |
The results obtained with MDF SAR were compared with the previous reported ones (table 2) using the Steigers Z test. The results of comparison between correlated correlations obtained with MDF SAR bi-varied model and previous reported results are in table 5.
Table 5. The models comparison results
|
Statistical parameter(s) |
Variables |
Value(s) |
|
|
1 |
2 |
||
|
r |
Insecticidal Activity |
Bi-varied MDF SAR |
0.9996 |
|
r |
Insecticidal Activity |
3W_PLS |
0.9325 |
|
r |
Insecticidal Activity |
S_PLS |
0.9556 |
|
r |
Bi-varied MDF SAR |
3W_PLS |
0.9312 |
|
r |
Bi-varied MDF SAR |
S_PLS |
0.9591 |
|
r |
S_PLS |
3W_PLS |
0.9698 |
|
Z, pZ(%) |
Bi-varied MDF SAR |
3W_PLS |
5.5, 2·10-5 |
|
Z, pZ(%) |
Bi-varied MDF SAR |
S_PLS |
5.1, 2·10-4 |
|
Z, pZ(%) |
S_PLS |
3W_PLS |
0.7, 25 |
Discussions
The mono-varied MDF SAR model of insecticidal activity of the eight neonicotinoid studied compounds use the iIDrSMg as molecular descriptor and gives us a probability of wrong model equal with 0.06%. The iIDrSMg descriptor take into consideration the geometric distance operator (g) and the atomic relative mass (M). Almost eighty-eight percent of the variation in insecticidal activity of studied neonicotinoid is explainable by its linear relation with iIDrSMg, being of the molecular geometry nature.
The bi-varied MDF SAR model use ImMdsEg and lIMMFQt descriptors, and is statistically significant (2·10-6 %). This model takes into consideration the geometric distance operator (g), the topological distance operator (t), the atomic electronegativity (E), as well as the partial charge, semi-empirical Extended Hückel model, Single Point approach (Q) in order to explain the insecticidal activity of the studied neonicotinoid. Ninety-nine percent of the variation in insecticidal activity of studied neonicotinoid is explainable by its linear relation with ImMdsEg and lIMMFQt descriptors.
The descriptor from the best mono-varied equation does not appear in the bi-varied best model. It can be says that is an expected observation, if we consider the bi-varied model as a refined of structure-activity dependency and the insecticidal activity are decomposed differently in two structural components. The absence of the best descriptor from the mono-varied model from the pair(s) of best bi-varied model demonstrates that it is no link between using of orthogonal descriptors (Principal and/or Dominant Component Analysis) and MDF SAR modeling.
All multiple linear regression models resulted from splitting the compounds in training and test set obtained appropriate r-squared values and were statistical significant, that demonstrate the validity of the MDF SAR model.
Our best model use only two molecular structure descriptors and compared with the previous reported model is significantly better (pZ = 2·10-5 comparing with 3W_PLS, pZ = 2·10-4 comparing with S_PLS). The MDF SAR model use only two structure descriptors (instead of three as was previous reported) and the statistical result is even better (squared correlation coefficient from 0.91 to 0.999 and cross-validation score from 0.8 to 0.998).
The obtained scores of correlation in bi-varied model make unpractical to search for better models with three or more molecular structure descriptors.
Even if the described structure-activity relationship methodology is a complex one and time consuming (takes about one or two days to complete the bi-varied model findings on a P3 server and client type machine) the results worth the effort.
Conclusions
The MDF SAR methodology is a better solution in predicting the insecticidal activity of the studied neonicotinoid compounds giving better results compared with the previous reported 3-way PLS method or standard PLD method.
The advantages represented by the simplicity of the best model and the easiness of the interpret it (it use just two molecular descriptors) and by the higher performances sustain the use of the using of the MDF SAR methodology in prediction of insecticidal activity of neonicotinoid compounds.
References