Pearson versus Spearman, Kendall's Tau Correlation Analysis on Structure-Activity Relationships of Biologic Active Compounds
Sorana-Daniela BOLBOACĂ1, Lorentz JÄNTSCHI 2
1Iuliu Haţieganu University of Medicine and Pharmacy, 13 Emil Isac, 400023 Cluj-Napoca, Romania; 2Technical University of Cluj-Napoca, 15 Constantin Daicoviciu, 400020 Cluj-Napoca, Romania
sbolboaca@umfcluj.ro, lori@academicdirect.org,
Abstract
A sample of sixty-seven pyrimidine derivatives with inhibitory activity on E. coli dihydrofolate reductase (DHFR) was studied by the use of molecular descriptors family on structure-activity relationships. Starting from the results obtained by applying of MDF-SAR methodology on pyrimidine derivatives and from the assumption that the measured activity (compounds inhibitory activity) of a biologically active compounds is a semi-quantitative outcome (can be related with the type of equipment used, the researchers, the chemical used, etc.), the abilities of Pearson, Spearman, Kendalls, and Gamma correlation coefficients in analysis of estimated toxicity were studied and are presented.
Keywords
Multiple linear regressions, Correlation coefficients, Molecular Descriptors Family on Structure-Activity Relationships (MDF-SAR)
Introduction
QSAR (Quantitative Structure-Activity Relationships) is an approach which is able to indicate for a given compound or a class of compounds which feature of structure characteristics is correlated with its activity [1]. In QSAR analysis were proposed several approaches for development. Simple and multiple linear regressions is one of the more successful techniques use by many researcher in construct of QSAR models [2]-[4].
Correlation coefficient is a simple statistical measure of relationship between one dependent and one or more than one independent variables and it is use as a measure of the statistical fit of a regression based model in QSAR [5]. Its squared value (the coefficient of determination) it is most frequently used parameter as a measure of the goodness-of-fit of the model [6]-[10].
A new approach of molecular descriptors family on structure-activity relationships (MDF-SAR) was developed [11], and proved its usefulness in estimation and prediction of: toxycity [12], [13], mutagenicity 12, antioxidant efficacy [14], antituberculotic activity [15], antimalarial activity [16], antiallergic activity [17], anti-HIV-1 potencies [18], inhibition activity on carbonic anhydrase II [19] and IV [20].
Several correlation coefficients based on different statistical hypothesis are known and most frequently used today: Pearson correlation coefficient, Spearman rank correlation coefficient and Spearman semi-quantitative correlation coefficient, Kendall tau-a, -b and -c correlation coefficients, Gamma correlation coefficient 5.
Starting from the results obtained by applying of MDF-SAR methodology on a sample of sixty-seven compounds and from the assumption that the measured activity (compounds inhibitory activity) of a biologically active compounds is a semi-quantitative outcome (can be related with the type of equipment used, the researchers, the chemical used), the abilities of Pearson, Spearman, Kendalls, and Gamma correlation coefficients in analysis of estimated toxicity were studied.
Multi-varied MDF-SAR model of pyrimidine derivatives
A sample of sixty-seven pyrimidine derivatives with inhibitory activity on E. coli dihydrofolate reductase (DHFR) was studied by the use of MDF-SAR methodology.
The set of pyrimidine derivatives (2,4-Diamino-5-(substituted-benzyl)-pyrimidine derivatives) with inhibitory activity on E. coli dihydrofolate reductase (DHFR) was previously studied by Ting-Lan Chiu & Sung-Sau So by the use of neural network approach [21].
By applying the MDF-SAR methodology on the sample of sixty-seven pyrimidine derivatives, a multi-varied model with four descriptors reveled to has good performances in prediction and estimation of inhibitory activity.
The multi-varied MDF-SAR model with four descriptors had the following equation:
Yest = 3.78 + 1.62·iImrKHt + 2.37·liMDWHg + 6.40·IsDrJQt - 8.52·10-2·LSPmEQg
Analyzing the MDF-SAR model with four descriptors it could be say that inhibitory activity consider compounds geometry (g) and topology (t), being related with the number of directly bonded hydrogens (H) of compounds and with the partial charge (Q) as atomic properties.
Statistical characteristics of the MDF-SAR model with four descriptors are in table 1 and 2.
Table 1. Statistical characteristics of the multi-varied MDF-SAR model with four descriptors
|
Characteristic (notation) |
Value |
|
Number of variable (v) |
4 |
|
Correlation coefficient (r) |
0.9517 |
|
95% Confidence Intervals for r (95% CIr) |
|
|
Squared correlation coefficient (r2) |
0.9058 |
|
Adjusted squared correlation coefficient (r2adj) |
0.8997 |
|
Standard error of estimated (sest) |
0.1919 |
|
Fisher parameter (Fest) |
149* |
|
Cross-validation leave-one-out (loo) score (r2cv-loo) |
0.8932 |
|
Fisher parameter for loo analysis (Fpred) |
130* |
|
Standard error for leave-one-out analysis (sloo) |
0.2044 |
|
Model stability (r2 - r2cv(loo)) |
0.0126 |
|
r2(iImrKHt, liMDWHg) |
0.2020 |
|
r2(iImrKHt, IsDrJQt) |
0.0047 |
|
r2(iImrKHt, LSPmEQg) |
0.1482 |
|
r2(liMDWHg, IsDrJQt) |
0.0003 |
|
r2(liMDWHg, LSPmEQg) |
0.0212 |
|
r2(IsDrJQt, LSPmEQg) |
0.0664 |
|
*p < 0.001 |
|
Table 2. Statistics of the regression MDF-SAR model with four descriptors
|
|
StdError |
t Stat |
95%CIcoefficient |
r(Ym,desc) |
|
Intercept |
0.1999 |
18.92* |
[3.38, 4.18] |
n.a. |
|
iImrKHt |
0.0709 |
22.85* |
[1.48, 1.76] |
0.4803 |
|
liMDWHg |
0.1500 |
15.81* |
[2.07, 2.67] |
0.0558 |
|
IsDrJQt |
1.4779 |
4.33* |
[3.45, 9.36] |
0.0336 |
|
LSPmEQg |
0.0182 |
-4.68* |
[-0.12, -0.12] |
0.0231 |
|
StdError = standard error; t Stat = Student tets parameter; 95% CIcoefficient = 95% confidence interval associated with regression coefficients; Ym = measured inhibitory activity; desc = molecular descriptor; * p < 0.001 |
||||
Graphical representation of the measured versus estimated by MDF-SAR model with four descriptors inhibitory activity is in figure 1.
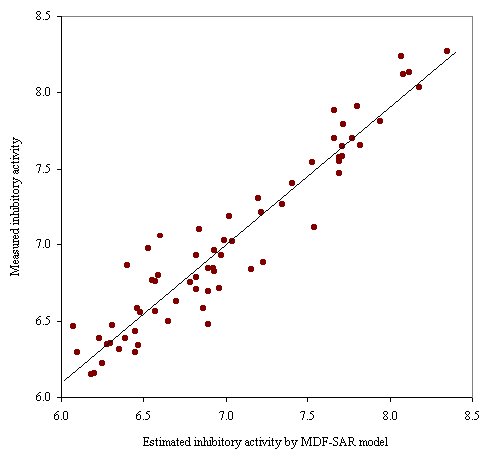
Figure 1. Plot of measured vs estimated by MDF-SAR inhibitory activity
Internal validation of the four-varied MDF SAR model with four descriptors was performed through splitting the whole set into training and test sets by applying of a randomization algorithm.
The coefficients for each model obtained in training sets, in conformity with the generic equation Yest = a0 + a1·iImrKHt + a2·liMDWHg + a3·IsDrJQt - a4·10-2·LSPmEQg, the number of compounds in training (Ntr) and test (Nts) sets, the correlation coefficient for training (rtr) and test (rts) sets with associated 95% confidence intervals (95%CIrtr and 95%CIrts), the Fisher parameter associated with training (Ftr) and test (Fts) sets, and the Fishers Z parameter of correlation coefficients comparison (Zrtr-rts) are in table 3.
Table 3. Statistics results on training versus test sets
|
a0 |
a1 |
a2 |
a3 |
a4 |
Ntr |
rtr |
95%CIrtr |
Ftr |
Nts |
rts |
95%CIts |
Fts |
Zrtr-rts |
|
3.93 |
1.61 |
2.43 |
6.56 |
-9.67·10-2 |
35 |
0.949 |
[0.899, 0.974] |
67* |
32 |
0.958 |
[0.916, 0.980] |
59* |
0.418 |
|
3.98 |
1.57 |
2.45 |
6.55 |
-7.52·10-2 |
36 |
0.951 |
[0.905, 0.975] |
73* |
31 |
0.951 |
[0.899, 0.976] |
61* |
0.000 |
|
3.84 |
1.55 |
2.15 |
9.08 |
-9.12·10-2 |
37 |
0.944 |
[0.893, 0.908] |
66* |
30 |
0.949 |
[0.895, 0.976] |
55* |
0.206 |
|
3.94 |
1.59 |
2.42 |
6.10 |
-8.18·10-2 |
38 |
0.951 |
[0.907, 0.974] |
78* |
29 |
0.947 |
[0.890, 0.975] |
50* |
0.144 |
|
3.91 |
1.56 |
2.25 |
8.22 |
-1.04·10-1 |
39 |
0.963 |
[0.931, 0.981] |
110* |
28 |
0.937 |
[0.867, 0.971] |
39* |
1.069 |
|
4.18 |
1.51 |
2.44 |
6.06 |
-7.22·10-2 |
40 |
0.956 |
[0.917, 0.975] |
92* |
27 |
0.936 |
[0.863, 0.971] |
35* |
0.721 |
|
3.76 |
1.63 |
2.32 |
7.35 |
-1.02·10-1 |
41 |
0.963 |
[0.931, 0.980] |
116* |
26 |
0.935 |
[0.858, 0.971] |
34* |
1.104 |
|
3.97 |
1.58 |
2.39 |
5.11 |
-9.36·10-2 |
42 |
0.956 |
[0.919, 0.976] |
99* |
25 |
0.954 |
[0.896, 0.980] |
34* |
0.115 |
|
3.64 |
1.64 |
2.30 |
7.00 |
-8.15·10-2 |
43 |
0.955 |
[0.917, 0.975] |
98* |
24 |
0.944 |
[0.873, 0.976] |
37* |
0.407 |
|
3.72 |
1.66 |
2.43 |
5.78 |
-8.12·10-2 |
44 |
0.938 |
[0.889, 0.966] |
72* |
23 |
0.964 |
[0.916, 0.985] |
54* |
1.030 |
|
3.59 |
1.64 |
2.25 |
4.94 |
-9.98·10-2 |
45 |
0.947 |
[0.904, 0.970] |
86* |
22 |
0.957 |
[0.898, 0.982] |
37* |
0.411 |
|
3.86 |
1.55 |
2.23 |
8.68 |
-8.86·10-2 |
46 |
0.940 |
[0.894 0.967] |
78* |
21 |
0.983 |
[0.958, 0.993] |
43* |
2.290* |
|
4.04 |
1.54 |
2.36 |
6.46 |
-7.31·10-2 |
47 |
0.949 |
[0.911, 0.972] |
96* |
20 |
0.963 |
[0.906, 0.985] |
34* |
0.538 |
|
3.63 |
1.63 |
2.24 |
4.27 |
-8.93·10-2 |
48 |
0.940 |
[0.895, 0.966] |
82* |
19 |
0.963 |
[0.904, 0.986] |
44* |
0.852 |
|
3.98 |
1.57 |
2.42 |
6.49 |
-8.59·10-2 |
49 |
0.946 |
[0.905, 0.969] |
93* |
18 |
0.960 |
[0.894, 0.985] |
36* |
0.535 |
|
3.77 |
1.61 |
2.32 |
6.37 |
-8.46·10-2 |
50 |
0.943 |
[0.902, 0.968] |
91* |
17 |
0.974 |
[0.927, 0.991] |
52* |
1.294 |
|
3.67 |
1.63 |
2.22 |
6.56 |
-1.01·10-1 |
51 |
0.954 |
[0.919, 0.973] |
115* |
16 |
0.950 |
[0.858, 0.983] |
17* |
0.126 |
|
3.81 |
1.61 |
2.39 |
6.87 |
-7.70·10-2 |
52 |
0.951 |
[0.916, 0.972] |
112* |
15 |
0.950 |
[0.853, 0.984] |
22* |
0.032 |
|
3.69 |
1.65 |
2.36 |
6.32 |
-8.21·10-2 |
53 |
0.953 |
[0.919, 0.972] |
118* |
14 |
0.956 |
[0.864, 0.986] |
17* |
0.128 |
|
3.97 |
1.56 |
2.40 |
6.16 |
-7.51·10-2 |
54 |
0.951 |
[0.916, 0.971] |
115* |
13 |
0.954 |
[0.851, 0.987] |
17* |
0.122 |
|
p > 0.05; * p < 0.01 |
|||||||||||||
Definitions, Formulas, Interpretations, PHP functions, and Results
A number of add notations were used in the study, as follows:
· Pearson product-moment correlation coefficient (named after Karl Pearson (1857 - 1936), a major contributor to the early development of statistics):
o rprs = the Pearson correlation coefficient;
o rPrs2 = the squared Pearson correlation coefficient;
o tPrs,df = the Student test parameter, and its significance pPrs,df at a significance level of 5% (where df = the degree of freedom);
· Spearmans rank correlation coefficient (named after Charles Spearman (1863 - 1945), English psychologist known for his work in statistics - factor analysis, and Spearman's rank correlation coefficient):
o rSpm = the Spearman rank correlation coefficient
o rSpm2 = the squared of Spearman rank correlation coefficient;
o tPrs,df = the Student test parameter, and its significance pSpm,df;
o rsQ2 = the squared of Spearman semi-Qantitative correlation coefficient;
o tsQ = the Student test parameter, and its significance psQ;
· Kendalls tau correlation coefficients (named after Maurice George Kendall (1907 - 1983), a prominent British statistician; published in monograph Rank Correlation in 1948);
o τKen,a = the Kendall tau-a correlation coefficient;
o τKen,a2 = the squared of Kendall tau-a correlation coefficient;
o ZKen,τa = the Z-test parameter of Kendall tau-a correlation coefficient, and its significance pKen,τa;
o τKen,b = the Kendall tau-b correlation coefficient;
o τKen,b2 = the squared of Kendall tau-b correlation coefficient;
o ZKen,τb = the Z-test parameter of Kendall tau-b, and its significance pKen,τb;
o τKen,c2 = the Kendall tau-c correlation coefficient;
o τKen,c2 = the squared of Kendall tau-c correlation coefficient;
o ZKen,τc = the Z-test parameter of Kendall tau-c, and its significance pKen,τc;
· Gamma correlation coefficient (also known as Goodman and Kruskal's gamma):
o Γ = the Gamma correlation coefficient;
o Γ2 = the squared of Gamma correlation coefficient;
o ZΓ = the Z-test parameter of Gamma correlation coefficient, and its significance pΓ.
A series of *.php programs which to facilitate the calculation and to display of above-described correlation coefficients and their statistics (Student-test and Z-test parameters and associated significances) were implemented and was use in order to reach the objective of study [22].
Pearson correlation coefficient
Definition: a measure the strength and direction of the linear relationship between two variables, describing the direction and degree to which one variable is linearly related to another.
Assumptions: both variable (variables Ym and Yest) are interval or ratio variables and are well approximated by a normal distribution, and their joint distribution is bivariate normal [23].
Formula
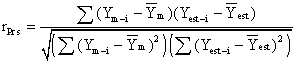
where Ym-i
is the value of the measured inhibitory activity for compound i (i = 1, 2,
,
67) ![]() is
the average of the measured inhibitory activity, Yest-i is the value
of the estimated inhibitory activity for compound i, and
is
the average of the measured inhibitory activity, Yest-i is the value
of the estimated inhibitory activity for compound i, and ![]() is the average of
the estimated inhibitory activity.
is the average of
the estimated inhibitory activity.
Interpretation
The Pearson correlation coefficient can take values from -1 to +1. A value of +1 show that the variables are perfectly linear related by an increasing relationship, a value of -1 show that the variables are perfectly linear related by an decreasing relationship, and a value of 0 show that the variables are not linear related by each other. There is considered a strong correlation if the correlation coefficient is greater than 0.8 and a weak correlation if the correlation coefficient is less than 0.5.
The coefficient of determination (or r squared) gives information about the proportion of variation in the dependent variable which might be considered as being associated with the variation in the independent variable.
Related statistics:
· The squared of Pearson correlation coefficient or Pearson coefficient of determination (rPrs2);
o Describe the proportion of variance in Ym that is related with linear variation of Yest;
o Can take values from 0 to 1.
Statistical test
Student t-test was used to determine if the value of Pearson correlation coefficient is statistically significant, at a significance level of 5%.
The null hypothesis vs. the alternative hypothesis was:
H0: rPrs = 0 (there is no correlation between the variables)
H1: rPrs < > 0 (variables are correlated)
For a significance level equal with 5%, a p-value associated to tPrs,df less than 0.05 means that there is evidence to reject the null hypothesis in favor of the alternative hypothesis. In other words there is a statistically significant linear relationship between the variables.
PHP implementation
In order to compute the statistics associated with Pearson correlation coefficient, three functions were implemented:
function coef_rk(&$y1,&$y2){
$my1=m1($y1);
$dy2=m2($y1,$y1)-$my1*$my1;
$mx1=m1($y2);
$mxy=m2($y2,$y1);
$m2x=$mx1*$mx1;
$mx2=m2($y2,$y2);
$dx2=$mx2-$m2x;
$r2=pow($mxy-$mx1*$my1,2)/($dx2*$dy2);
return $r2;
}
function t_p($n,$k,$r){
return $r*pow($n-$k-1,0.5)/pow(1-pow($r,2),0.5);
}
function p_t($t,$df){
$p = $df/2;
$x = 0.5+0.5*$t/pow(pow($t,2)+$df,0.5);
$beta_gam = exp( -logBeta($p, $p) + $p * log($x) + $p * log(1.0 - $x) );
return (2.0 * $beta_gam * betaFraction(1.0 - $x, $p, $p) / $p);
}
The statistics of Pearson correlation coefficients are computed as follows:
· Pearson correlation coefficient:
$r_pe = coef_rk($cmp[0],$cmp[1]);
where $cmp[0] is the measured inhibitory activity (Ym), and $cmp[1] is the estimated by MDF-SAR model with four descriptor inhibitory activity (Yest).
· t Student parameter:
$t_pe = t_p($n,1,pow($r_pe,0.5));
· Significance of t Student parameter
$p_pe = p_t($t_pe,$n-2);
Results:
rPrs2 = 0.9058
tPrs,1 = 24.99 (1)
pPrs,1 = 4.74·10-33 %
Spearmans rank correlation coefficient
Definition: a non-parametric measure of correlation between variable which assess how well an arbitrary monotonic function could describe the relationship between two variables, without making any assumptions about the frequency distribution of the variables. Frequently the Greek letter ρ (rho) is use to abbreviate the Spearman correlation coefficient.
Spearmans rank correlation is satisfactory for testing the null hypothesis of no relationship, but is difficult to interpret as a measure of the strength of the relationship [24].
Assumptions:
· Does not required any assumptions about the frequency distribution of the variables;
· Does not required the assumption that the relationship between variable is linear;
· Does not required the variable to be measured on interval or ration scale.
Formula
In order to compute the Spearman rank correlation coefficient, the two variables (Ym, respectively Yest) were converted to ranks (see table 4 for exemplification). For each measured and estimated inhibitory activity a rank was assigned (RankYm - for measure inhibitory activity, RankYest - for estimated by MDF-SAR model inhibitory activity) according with the position of value into a sort serried of values.
In assignment of rank process, the lowest value had the lowest rank. When there are two equal values for two different compounds (for measured and/or estimated inhibitory activity), the associated rank had equal values and was calculated as means of corresponding ranks. For example, the compounds abbreviated as c_52 and c_59 have the same measured inhibitory activity (6.45, see table 4). The rank associated with these values is equal with 13.5 (is the average between the rank for c_52 - 13 and the rank of c_59 - 14).
Table 4. Compounds abbreviation, measured and estimated activity and associated ranks
|
Abb. |
Ym |
RankYm |
|
Yest |
RankYest |
|
Abb. |
Ym |
RankYm |
|
Yest |
RankYest |
|
c_64 |
6.07 |
1 |
0 |
6.4626 |
13 |
|
c_32 |
6.92 |
35 |
0 |
6.8423 |
32 |
|
c_65 |
6.10 |
2 |
0 |
6.2948 |
5 |
|
c_66 |
6.93 |
36.5 |
5 |
6.8225 |
30 |
|
c_67 |
6.18 |
3 |
0 |
6.1479 |
1 |
|
c_36 |
6.93 |
36.5 |
6.9609 |
38 |
|
|
c_54 |
6.20 |
4 |
0 |
6.1595 |
2 |
|
c_40 |
6.96 |
38 |
0 |
6.7150 |
24 |
|
c_37 |
6.23 |
5 |
0 |
6.3859 |
10 |
|
c_17 |
6.97 |
39 |
0 |
6.9298 |
36 |
|
c_48 |
6.25 |
6 |
0 |
6.2254 |
3 |
|
c_45 |
6.99 |
40 |
0 |
7.0283 |
41 |
|
c_31 |
6.28 |
7 |
0 |
6.3483 |
8 |
|
c_41 |
7.02 |
41 |
0 |
7.1919 |
45 |
|
c_49 |
6.30 |
8 |
0 |
6.3528 |
9 |
|
c_15 |
7.04 |
42 |
0 |
7.0225 |
40 |
|
c_10 |
6.31 |
9 |
0 |
6.4703 |
14 |
|
c_28 |
7.16 |
43 |
0 |
6.8355 |
31 |
|
c_56 |
6.35 |
10 |
0 |
6.3149 |
6 |
|
c_09 |
7.20 |
44 |
0 |
7.3115 |
48 |
|
c_47 |
6.39 |
11 |
0 |
6.3866 |
11 |
|
c_18 |
7.22 |
45 |
0 |
7.2156 |
46 |
|
c_53 |
6.40 |
12 |
0 |
6.8614 |
34 |
|
c_43 |
7.23 |
46 |
0 |
6.8855 |
35 |
|
c_52 |
6.45 |
13.5 |
1 |
6.2913 |
4 |
|
c_29 |
7.35 |
47 |
0 |
7.2724 |
47 |
|
c_59 |
6.45 |
13.5 |
6.4336 |
12 |
|
c_14 |
7.41 |
48 |
0 |
7.4072 |
49 |
|
|
c_16 |
6.46 |
15 |
0 |
6.5851 |
20 |
|
c_24 |
7.53 |
49 |
0 |
7.5476 |
51 |
|
c_34 |
6.47 |
16 |
0 |
6.3422 |
7 |
|
c_22 |
7.54 |
50 |
0 |
7.1218 |
44 |
|
c_58 |
6.48 |
17 |
0 |
6.5536 |
17 |
|
c_26 |
7.66 |
51.5 |
6 |
7.7002 |
57 |
|
c_35 |
6.53 |
18 |
0 |
6.9755 |
39 |
|
c_08 |
7.66 |
51.5 |
7.8841 |
61 |
|
|
c_42 |
6.55 |
19 |
0 |
6.7654 |
27 |
|
c_27 |
7.69 |
54 |
7 |
7.4715 |
50 |
|
c_30 |
6.57 |
20.5 |
2 |
6.5625 |
18 |
|
c_13 |
7.69 |
54 |
7.5489 |
52 |
|
|
c_61 |
6.57 |
20.5 |
6.7594 |
26 |
|
c_12 |
7.69 |
54 |
7.5793 |
53 |
||
|
c_33 |
6.59 |
22 |
0 |
6.8010 |
29 |
|
c_04 |
7.71 |
56.5 |
8 |
7.5841 |
54 |
|
c_51 |
6.60 |
23 |
0 |
7.0616 |
42 |
|
c_11 |
7.71 |
56.5 |
7.6497 |
55 |
|
|
c_39 |
6.65 |
24 |
0 |
6.4993 |
16 |
|
c_19 |
7.72 |
58 |
0 |
7.7915 |
59 |
|
c_38 |
6.70 |
25 |
0 |
6.6297 |
21 |
|
c_23 |
7.77 |
59 |
0 |
7.7014 |
58 |
|
c_57 |
6.78 |
26 |
0 |
6.7552 |
25 |
|
c_25 |
7.80 |
60 |
0 |
7.9130 |
62 |
|
c_60 |
6.82 |
28 |
3
|
6.7091 |
23 |
|
c_01 |
7.82 |
61 |
0 |
7.6576 |
56 |
|
c_44 |
6.82 |
28 |
6.7847 |
28 |
|
c_21 |
7.94 |
62 |
0 |
7.8130 |
60 |
|
|
c_55 |
6.82 |
28 |
6.9318 |
37 |
|
c_06 |
8.07 |
63 |
0 |
8.2391 |
66 |
|
|
c_20 |
6.84 |
30 |
0 |
7.1067 |
43 |
|
c_03 |
8.08 |
64 |
0 |
8.1224 |
64 |
|
c_46 |
6.86 |
31 |
0 |
6.5813 |
19 |
|
c_07 |
8.12 |
65 |
0 |
8.1353 |
65 |
|
c_50 |
6.89 |
33 |
4
|
6.4794 |
15 |
|
c_05 |
8.18 |
66 |
0 |
8.0372 |
63 |
|
c_62 |
6.89 |
33 |
6.6942 |
22 |
|
c_02 |
8.35 |
67 |
0 |
8.2702 |
67 |
|
|
c_63 |
6.89 |
33 |
6.8475 |
33 |
|
|
|
|
|
|
|
The method of rank assignment for more then two equal values of measured and/or estimated inhibitory activity is the same as for two equal values. If there are an odd number of compounds which have the same measured value (see compounds c_60, c_44, and c_55 from table 2) then the rank will be an integer ((27+28+29)/3 = 28, see the rank for c_60, c_44, and c_55).
In studied example, there are equal values for measured activity: five situations of two equal values (c_52-c_59, c_30-c_61, c_66-c_36, c_26-c_08, and c_04-c_11), and three situations of three equal values (c_60-c_44-c_55, c_50-c_62-c_63, and c_27-c_13-c_12).
By conversion of the measured and estimated inhibitory activity to ranks, the distribution of ranks does not depend on the distribution of measured, respectively estimated inhibitory activity.
The formula for calculation of the Spearman rank correlation coefficient is:
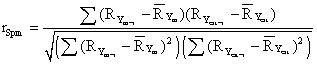
where RYm-i
is the rank of the measured inhibitory activity for compound i, ![]() is the
average of the measured inhibitory activity, RYest-i is the rank of
the estimated by MDF-SAR inhibitory activity for compound i, and
is the
average of the measured inhibitory activity, RYest-i is the rank of
the estimated by MDF-SAR inhibitory activity for compound i, and ![]() is the
average of the estimated inhibitory activity.
is the
average of the estimated inhibitory activity.
The simple formula for rSpm is based on the difference between each pairs of ranks:
![]()
where D is the differences between each pair of ranks (e.g. D = RYm-1 - RYest-1) and n is the volume of the sample.
The formula of the Spearman semi-quantitative method is:
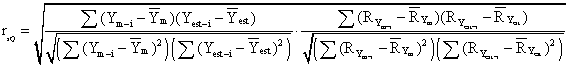
Interpretation
· Identical with Pearson correlation coefficient.
Related statistics:
· rSpm2 = the squared of Spearman rank correlation coefficient;
· rsQ2 = the squared of semi-quantitative correlation coefficient.
Statistical significance:
· Compute by the use of a permutation test (a statistical test in which the reference distribution is obtained by permuting the observed data points across all possible outcomes, given a set of conditions consistent with the null hypothesis);
· Comparing the observed rSpm with published tables for different levels of significance (eg. 0.05, 0.01 ). It is a simple solution when the researchers want to know the significance within a certain range or less than a certain value;
· Tested by applying the Student t-test (for sample sizes > 20): the method used in this study.
The null hypothesis vs. the alternative hypothesis for Spearman rank correlation coefficient was:
H0: rSpm = 0 (there is no correlation between the ranked pairs)
H1: rSpm < > 0 (ranked pairs are correlated)
The null hypothesis vs. the alternative hypothesis for semi-quantitative correlation coefficient was:
H0: rsQ = 0 (there is no correlation between the ranked pairs)
H1: rsQ < > 0 (ranked pairs are correlated)
PHP implementation
The formulas for Spearman and respectively semi-quantitative correlation coefficients used two defined above functions (t_p and respectively p_t). The Spearman rank correlation coefficient used the coef_rk function defined as:
function coef_rk(&$y1,&$y2){
$my1=m1($y1);
$dy2=m2($y1,$y1)-$my1*$my1;
$mx1=m1($y2);
$mxy=m2($y2,$y1);
$m2x=$mx1*$mx1;
$mx2=m2($y2,$y2);
$dx2=$mx2-$m2x;
$r2=pow($mxy-$mx1*$my1,2)/($dx2*$dy2);
return $r2;
}
where
function m1(&$v){
$rez=0;
$n=count($v);
for($i=1;$i<$n;$i++)
$rez+=$v[$i];
return $rez/($n-1);
}
function m2(&$v,&$u){
$rez=0;
$n=count($v);
for($i=1;$i<$n;$i++)
$rez+=$v[$i]*$u[$i];
return $rez/($n-1);
}
Spearman correlation coefficient
The statistics of Spearman rank correlation coefficients are computed as follows:
· Spearman correlation coefficient:
$r_sp = coef_rk($poz[0],$poz[1]);
where $poz[0] is the position on sort series of measured inhibitory activity, and $poz[1] is the position on sort serried of estimated inhibitory activity by MDF-SAR model with four descriptor.
· t Student parameter:
$t_sp = t_p($n,1,pow($r_sp,0.5));
· Significance of t Student parameter
$p_sp = p_t($t_sp,$n-2);
Semi-quantitative correlation coefficient
The statistics of semi-quantitative correlation coefficients are computed as follows:
· Semi-quantitative correlation coefficient:
$r_sq = pow($r_pe*$r_sp,0.5);
· t Student parameter:
$t_sq = t_p($n,1,pow($r_sq,0.5));
· Significance of t Student parameter
$p_sq = p_t($t_sq,$n-2);
Results:
rSpr2 = 0.8606
tSpm,1 = 20.03 (2)
pSpm,1 = 1.62·10-29
rsQ2 = 0.8829
tsQ = 22.14 (3)
psQ = 5.57·10-32
Kendalls rank correlation coefficients
Definition
Kendall-tau is a non-parametric correlation coefficient that can be used to assess and test correlations between non-interval scaled ordinal variables. Frequently the Greek letter τ (tau), is use to abbreviate the Kendall tau correlation coefficient.
The Kendall tau correlation coefficient is considered to be equivalent to the Spearman rank correlation coefficient. While Spearman rank correlation coefficient is like the Pearson correlation coefficient but computed from ranks, the Kendall tau correlation rather represents a probability.
There are three Kendalls tau correlation coefficient known as tau-a, tau-b, and tau-c.
Formula
Let (Ym-i, Yest-i) and (Ym-j, Yest-j) be the pair of measured and estimated inhibitory activity. If Ym-j - Ym-i and Yest-j - Yest-i, where i < j have the same sign the pair is concordant, if have opposite signs the pair is discordant.
In a sample of n observations it can be found n(n-1)/2 pairs corresponding to choices 1 ≤ i < j ≤ n.
The formulas of Kendalls tau correlation coefficients are as follows:
· Kendall tau-a correlation coefficient (τKen,a):
τKen,a = (C-D)/[n(n-1)/2]
· Kendall tau-b correlation coefficient (τKen,b):
τKen,b = (C-D)/Ö[(n(n-1)/2-t)(n(n-1)/2-u)]
where t is the number of tied Ym values and u is the number of tied Yest values.
· Kendall tau-c correlation coefficient (τKen,c):
τKen,c = 2(C-D)/n2
Interpretation:
· If the agreement between the two rankings is perfect and the two rankings are the same, the coefficient has value 1.
· If the disagreement between the two rankings is perfect and one ranking is the reverse of the other, the coefficient has value -1.
· For all other arrangements the value lies between -1 and 1, and increasing values imply increasing agreement between the rankings.
· If the rankings are independent, the coefficient has value 0.
Related statistics:
· τKen,a2 = the squared of Kendall tau-a correlation coefficient;
· τKen,b2 = the squared of Kendall tau-b correlation coefficient;
· τKen,c2 = the squared of Kendall tau-c correlation coefficient.
Statistical significance:
Statistical significance of the Kendalls tau correlation coefficient is testes by the Z-test, at a significance level of 5%. The null hypothesis vs. the alternative hypothesis for Kendalls tau correlation coefficients was:
· Kendall tau-a correlation coefficient:
H0: τKen,a = 0 (there is no correlation between the two variables)
H1: τKen,a < > 0 (the two variables are correlated)
· Kendall tau-b correlation coefficient:
H0: τKen,b = 0 (there is no correlation between the two variables)
H1: τKen,b < > 0 (the two variables are correlated)
· Kendall tau-b correlation coefficient:
H0: τKen,c = 0 (there is no correlation between the two variables)
H1: τKen,c < > 0 (the two variables are correlated)
PHP implementation
Kendall function was implemented in order to calculate the Kendalls tau correlation coefficients:
function Kendall(&$cmp){
$n = count($cmp[0]);
$pz = 0;
if(!is_numeric($cmp[0][0])) $pz = 1;
$C = 0;
$D = 0;
$E = 0;
for($i=$pz;$i<$n-1;$i++)
for($j=$i+1;$j<$n;$j++){
$sgx = 0;
$sgy = 0;
if($cmp[0][$i]>$cmp[0][$j]) $sgx = 1;
if($cmp[0][$i]<$cmp[0][$j]) $sgx = -1;
if($cmp[1][$i]>$cmp[1][$j]) $sgy = 1;
if($cmp[1][$i]<$cmp[1][$j]) $sgy = -1;
if($sgx*$sgy>0) $C++;
if($sgx*$sgy<0) $D++;
if($sgx*$sgy==0) $E++;
if($sgx==0)$tied_x[$i][]=$j;
if($sgy==0)$tied_y[$i][]=$j;
}
$t1 = 0;
$u1 = 0;
$vt = 0;
$vu = 0;
$v2t = 0;
$v2u = 0;
if(isset($tied_x))
if(is_array($tied_x)){
foreach($tied_x as $vx){
$nt = count($vx)+1;
$t1 += $nt*($nt-1);
$vt += $nt*($nt-1)*(2*$nt+5);
$v2t += $nt*($nt-1)*($nt-2);
}
}
if(isset($tied_y))
if(is_array($tied_y)){
foreach($tied_y as $vy){
$nu = count($vy)+1;
$u1 += $nu*($nu-1);
$vu += $nu*($nu-1)*(2*$nu+5);
$v2u += $nu*($nu-1)*($nu-2);
}
}
$v1 = $t1*$u1;
$t1 /= 2;
$u1 /= 2;
$v2 = $v2t*$v2u;
$S = $C - $D;
$n = $n - $pz;
$cn2 = $n*($n-1)/2;
$tau_a2 = pow($S,2)/pow($cn2,2);
$v_tau_a = $cn2*(2*$n+5)/9;
$z_tau_a = $S/pow($v_tau_a,0.5);
$T = ($cn2-$t1)*($cn2-$u1);
$tau_b2 = pow($S,2)/$T;
$vT0 = $v_tau_a - ($vt + $vu)/18;
$vT1 = $v1/(4*$cn2);
$vT2 = $v2/(18*$cn2*($n-2));
$v_tau_b = pow($vT0 + $vT1 + $vT2 , 0.5);
$z_tau_b = $S/$v_tau_b;
$gamma = pow(($C - $D)/($C + $D),2);
$v_gamma = (2*$n+5)/9.0/$cn2;
$z_gamma = $gamma/pow($v_gamma,0.5);
$tau_c2 = 4*pow($S,2)/pow($n,4);
$z_tau_c = $z_tau_b*($n-1)/$n;
return array( $tau_a2, $z_tau_a, $tau_b2, $z_tau_b, $tau_c2, $z_tau_c, $gamma, $z_gamma );
}
where C is the number of concordant pairs (C = (<, <) or (>, >)), D is the number of discordant pairs (D = (<, >) or (>, <)), and E is the number of equal pairs (E = (=, .) or (., =)).
Results:
· Kendall's τa correlation coefficient and associated statistics:
τKen,a2 = 0.6129
ZKen,τa = 9.37 (4)
pKen,τa = 7.44·10-21
· Kendall's τb correlation coefficient and associated statistics:
τKen,b2 = 0.6177
ZKen,τb = 9.37 (5)
pKen,τb = 7.26·10-21
· Kendall's τc correlation coefficient and associated statistics:
τKen,c2 = 0.5948
ZKen,τc = 9.23 (6)
pKen,τc = 2.70·10-20
Gamma correlation coefficient
Definition
The Gamma correlation coefficient (Γ, gamma) is a measure of association between variables that comparing with Kendalls tau correlation coefficients is more resistant to tied data [25], being preferable to Spearman rank or Kendall tau when data contain many tied observations [26].
Formula
The formula for Gamma correlation coefficient is:
Γ = (C-D)/(C+D)
where the significance of C and D were described above.
Interpretation:
· In the same manner as the Kendall tau correlation coefficient.
Related statistics:
· Γ2 = the squared of Gamma correlation coefficient.
Statistical significance:
Statistical significance of Gamma correlation coefficient was tested by the Z-test, at a significance level of 5%. The null hypothesis vs. the alternative hypothesis for Gamma correlation coefficients was:
H0: Γ = 0 (there is no correlation between the two variables)
H1: Γ < > 0 (the two variables are correlated).
PHP implementation
The function which computes the Gamma correlation coefficient was presented at Kendalls tau correlation coefficient, in PHP implementation chapter.
Results:
Γ2 = 0.6208
ZΓ = 7.43 (7)
pΓ = 1.11·10-13
Conclusions
All seven computational methods used to evaluate the correlation between measured and estimated by MDF-SAR model inhibitory activity are statistically significant (p-value always less than 0.0001, correlation coefficients always greater than 0.5).
More research on other classes of biologic active compounds may reveal whether it is appropriate to analyze the MDF-SAR models using the Pearson correlation coefficient or other correlation coefficients (Spearman rank, Kendalls tau, or Gamma correlation coefficient).
Acknowledgement
Research was partly supported by UEFISCSU Romania through the project ET46/2006.
References